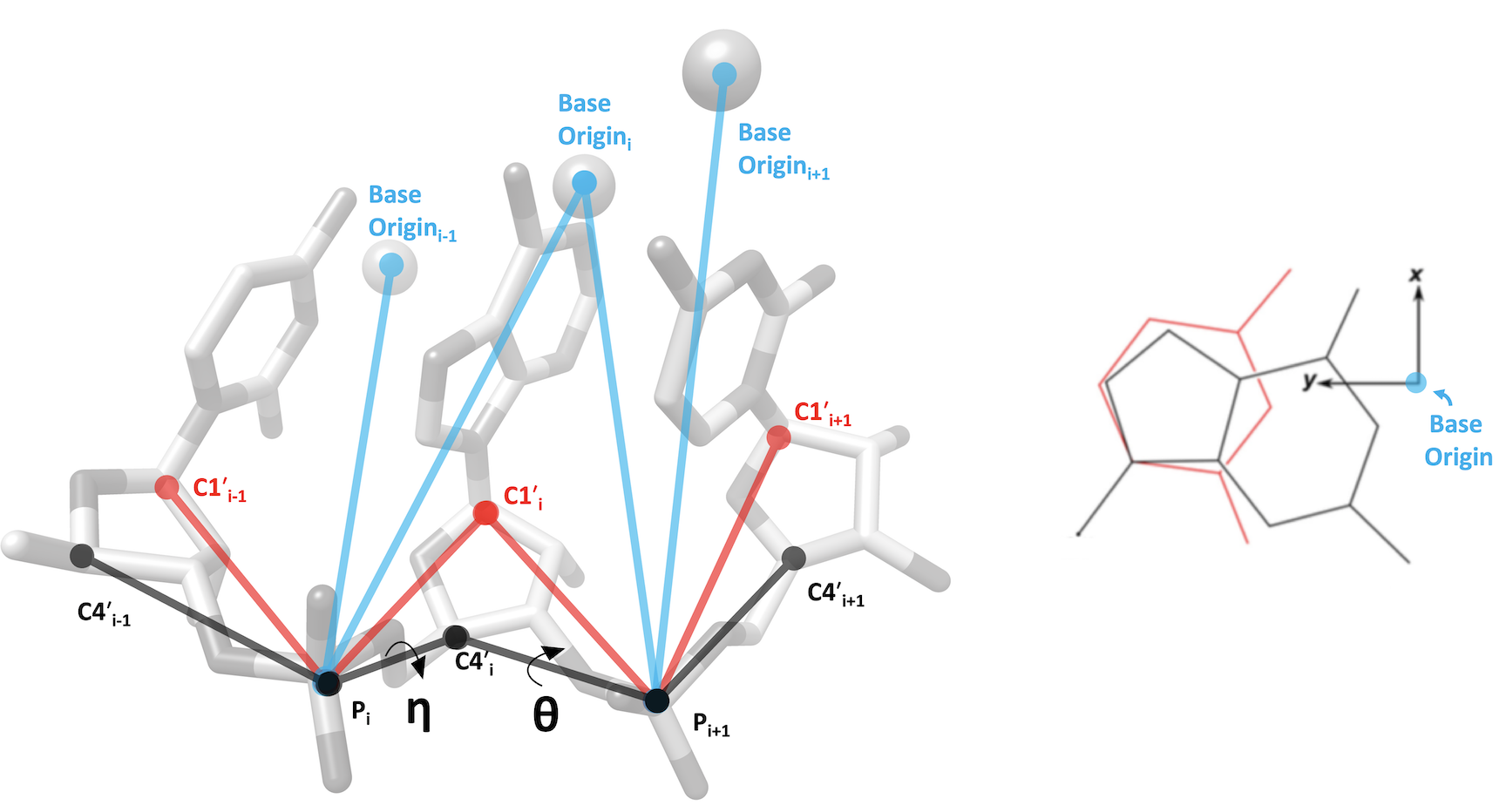
RNA Pseudotorsion Plots for PDB entry 4dv0
Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G Deposited 2012-02-22 X-RAY DIFFRACTION resol 3.853 Å
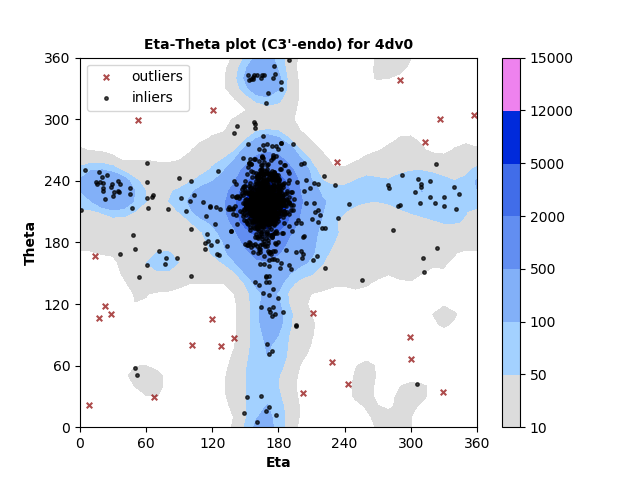
1326 C3'-endo RNA residues with 24 outliers ( 1.81 % ) from Eta-Theta plot
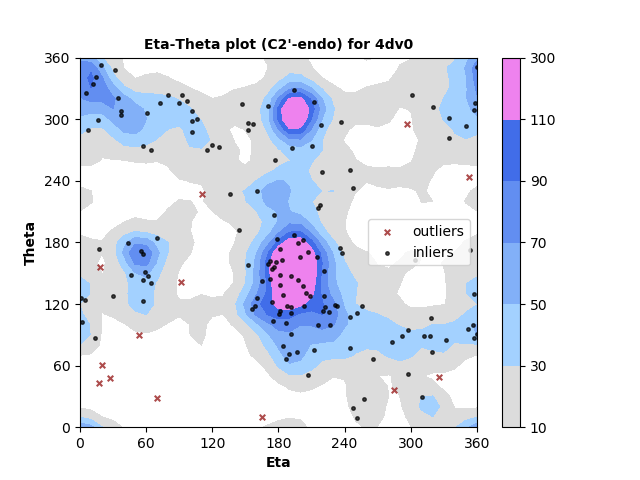
163 C2'-endo RNA residues with 13 outliers ( 7.98 % ) from Eta-Theta plot

1326 C3'-endo RNA residues with 17 outliers ( 1.28 % ) from Eta'-Theta' plot
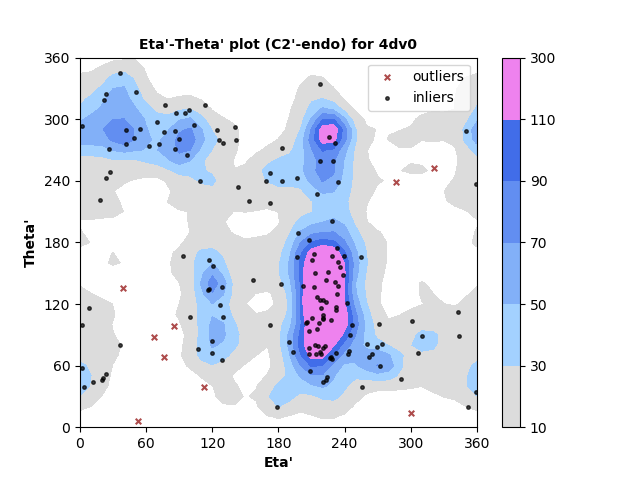
163 C2'-endo RNA residues with 9 outliers ( 5.52 % ) from Eta'-Theta' plot
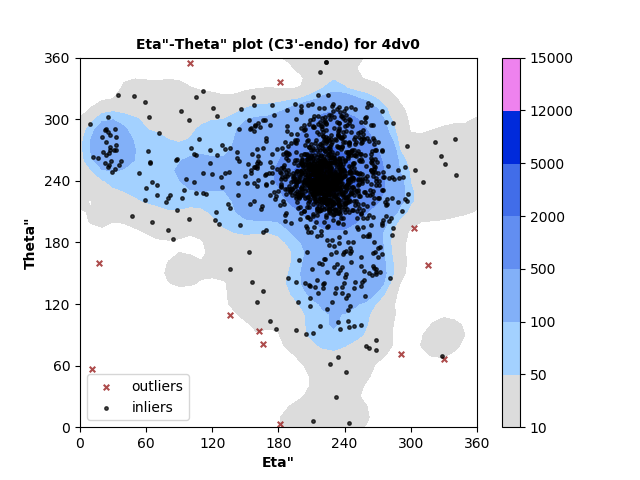
1326 C3'-endo RNA residues with 12 outliers ( 0.9 % ) from Eta"-Theta" plot
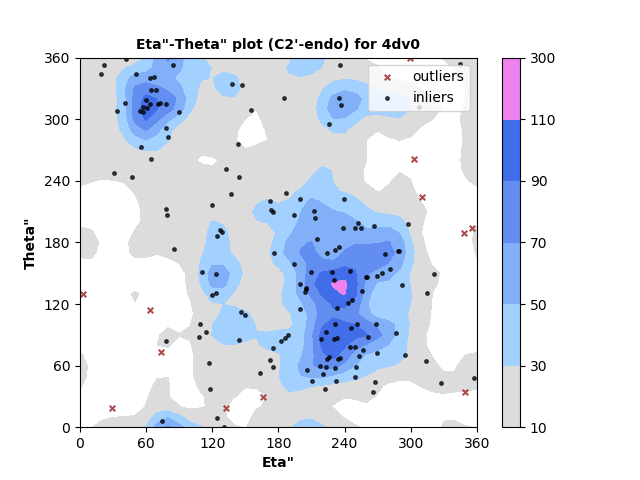
163 C2'-endo RNA residues with 12 outliers ( 7.36 % ) from Eta"-Theta" plot
List of Outliers
| Model Number | Chain | Residue Type | Residue Identifier | Eta | Theta | Eta' | Theta' | Eta'' | Theta'' | Sugar Pucker | Suiteness |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | C | 352 | 102.0 | 80.3 | 94.3 | 66.1 | 99.9 | 355.0 | C3'endo | 0.0 |
| 1 | A | A | 353 | 17.5 | 106.3 | 8.7 | 111.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 412 | 299.1 | 88.2 | 306.4 | 75.1 | 291.5 | 71.3 | C3'endo | 0.0 |
| 1 | A | G | 413 | 140.1 | 86.9 | 143.0 | 98.3 | --- | --- | C3'endo | 0.348 |
| 1 | A | G | 530 | 202.1 | 33.7 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 532 | 312.7 | 277.4 | --- | --- | --- | --- | C3'endo | 0.024 |
| 1 | A | U | 723 | 53.0 | 299.4 | 63.3 | 294.9 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 754 | 120.2 | 309.2 | 107.5 | 313.3 | --- | --- | C3'endo | 0.749 |
| 1 | A | A | 816 | 67.2 | 29.6 | --- | --- | --- | --- | C3'endo | 0.591 |
| 1 | A | U | 839 | 8.0 | 22.0 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 841 | 357.1 | 304.3 | 6.8 | 301.7 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1005 | 232.8 | 258.0 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1054 | 22.9 | 118.4 | 8.2 | 107.8 | 11.1 | 57.1 | C3'endo | 0.0 |
| 1 | A | U | 1125 | 28.4 | 109.8 | 18.7 | 119.9 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1126 | 326.7 | 300.4 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1200 | 228.8 | 63.3 | 229.0 | 36.0 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1212 | 299.9 | 66.5 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1278 | 242.8 | 42.5 | 247.7 | 52.3 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1300 | 329.7 | 33.8 | 298.6 | 73.4 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1398 | 128.3 | 79.1 | --- | --- | --- | --- | C3'endo | 0.01 |
| 1 | A | A | 1446 | 13.5 | 166.7 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1493 | 211.7 | 111.0 | 238.0 | 137.8 | 315.7 | 157.5 | C3'endo | 0.0 |
| 1 | A | G | 1530 | 119.9 | 105.4 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 722 | --- | --- | 128.5 | 144.1 | 166.3 | 81.0 | C3'endo | 0.785 |
| 1 | A | G | 1347 | --- | --- | 256.0 | 66.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1362 | --- | --- | 130.3 | 351.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1443 | --- | --- | 33.9 | 51.9 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 6 | --- | --- | --- | --- | 136.3 | 109.5 | C3'endo | 0.0 |
| 1 | A | U | 368 | 290.1 | 338.1 | --- | --- | 330.0 | 66.3 | C3'endo | 0.176 |
| 1 | A | A | 665 | --- | --- | --- | --- | 17.7 | 159.5 | C3'endo | 0.0 |
| 1 | A | A | 1377 | --- | --- | --- | --- | 162.7 | 93.2 | C3'endo | 0.671 |
| 1 | A | A | 1492 | --- | --- | --- | --- | 181.3 | 336.5 | C3'endo | 0.492 |
| 1 | A | G | 1505 | --- | --- | --- | --- | 181.4 | 3.0 | C3'endo | 0.0 |
| 1 | A | G | 1520 | --- | --- | --- | --- | 303.1 | 193.6 | C3'endo | 0.0 |
| 1 | A | A | 130 | 353.1 | 243.6 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 367 | 111.1 | 227.6 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 451 | 18.4 | 155.9 | 85.3 | 99.1 | --- | --- | C2'endo | 0.289 |
| 1 | A | A | 815 | 69.6 | 28.1 | 52.8 | 5.7 | --- | --- | C2'endo | 0.0 |
| 1 | A | C | 817 | 91.3 | 141.6 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 820 | 165.5 | 10.4 | --- | --- | --- | --- | C2'endo | 0.277 |
| 1 | A | A | 974 | 53.8 | 90.1 | 112.1 | 39.5 | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 992 | 17.2 | 43.0 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | G | 1094 | 20.1 | 60.2 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | C | 1129 | 284.6 | 36.5 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | C | 1397 | 325.6 | 48.7 | --- | --- | --- | --- | C2'endo | 0.586 |
| 1 | A | A | 8 | --- | --- | 286.5 | 238.8 | 303.3 | 261.1 | C2'endo | 0.0 |
| 1 | A | A | 50 | --- | --- | 67.5 | 87.5 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 60 | --- | --- | 38.7 | 135.4 | --- | --- | C2'endo | 0.199 |
| 1 | A | U | 190E | --- | --- | 320.8 | 252.6 | 310.2 | 224.0 | C2'endo | 0.0 |
| 1 | A | U | 1196 | --- | --- | 300.4 | 13.5 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1363 | --- | --- | 75.8 | 68.1 | 132.1 | 19.0 | C2'endo | 0.109 |
| 1 | A | A | 51 | 27.5 | 48.1 | --- | --- | 349.5 | 34.1 | C2'endo | 0.0 |
| 1 | A | U | 65 | --- | --- | --- | --- | 2.9 | 129.3 | C2'endo | 0.22 |
| 1 | A | G | 129A | --- | --- | --- | --- | 28.9 | 18.7 | C2'endo | 0.0 |
| 1 | A | A | 533 | 296.2 | 295.7 | --- | --- | 348.8 | 189.0 | C2'endo | 0.0 |
| 1 | A | U | 561 | --- | --- | --- | --- | 63.9 | 114.4 | C2'endo | 0.134 |
| 1 | A | U | 870 | --- | --- | --- | --- | 165.7 | 28.9 | C2'endo | 0.622 |
| 1 | A | U | 960 | --- | --- | --- | --- | 299.6 | 359.7 | C2'endo | 0.0 |
| 1 | A | A | 1157 | --- | --- | --- | --- | 73.5 | 72.9 | C2'endo | 0.0 |
| 1 | A | A | 1183 | --- | --- | --- | --- | 355.3 | 194.5 | C2'endo | 0.0 |