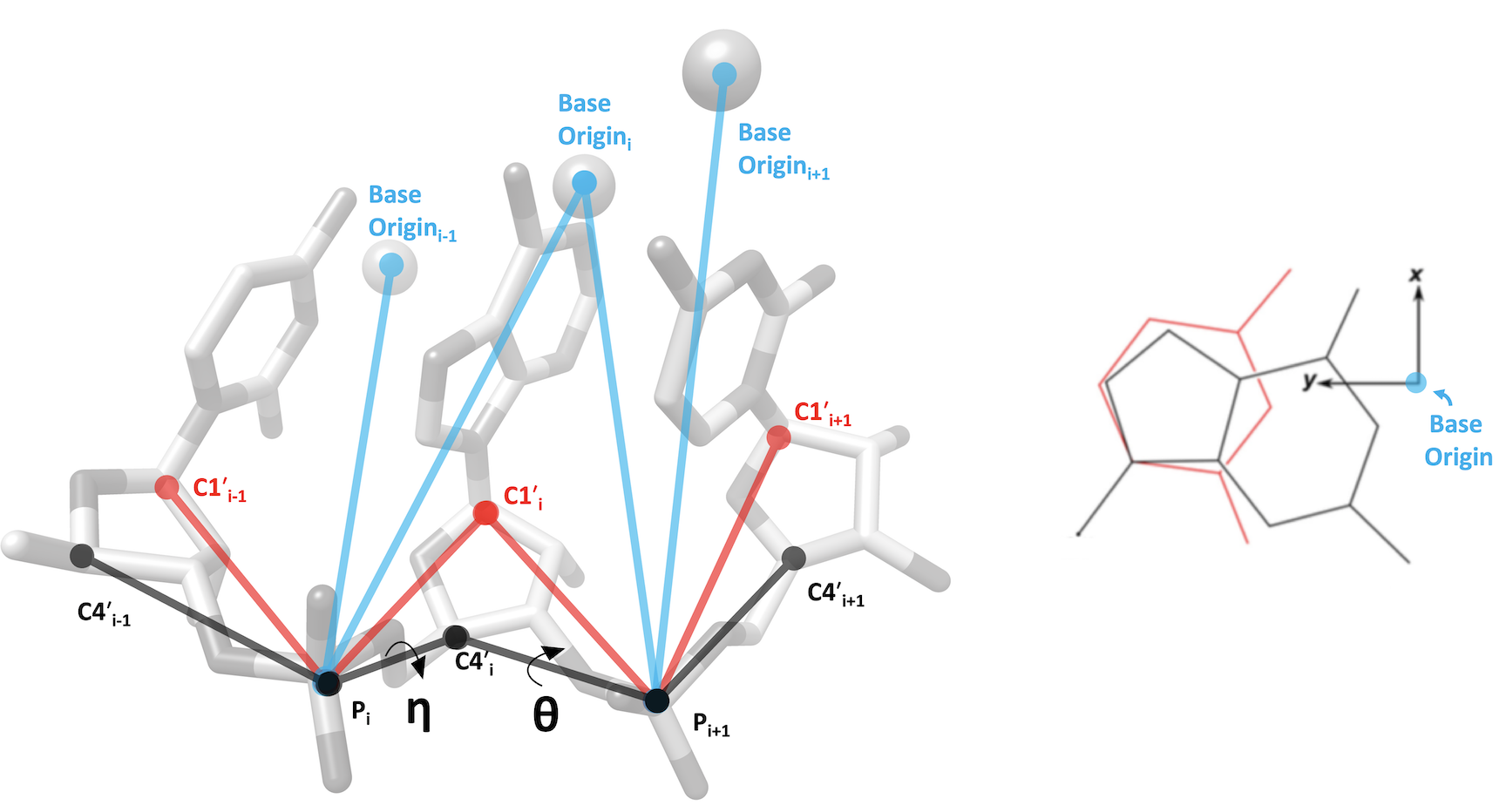
RNA Pseudotorsion Plots for PDB entry 4lf7
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus Deposited 2013-06-26 X-RAY DIFFRACTION resol 3.1484 Å
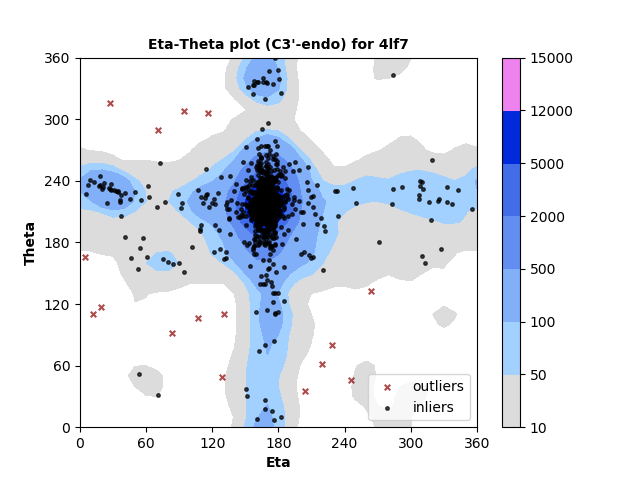
1310 C3'-endo RNA residues with 16 outliers ( 1.22 % ) from Eta-Theta plot
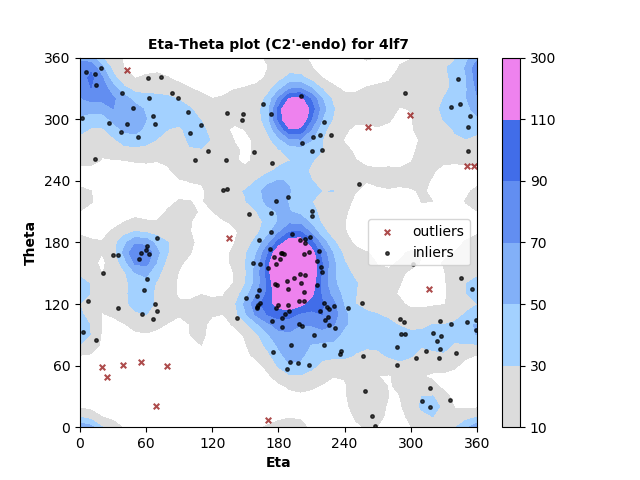
193 C2'-endo RNA residues with 14 outliers ( 7.25 % ) from Eta-Theta plot
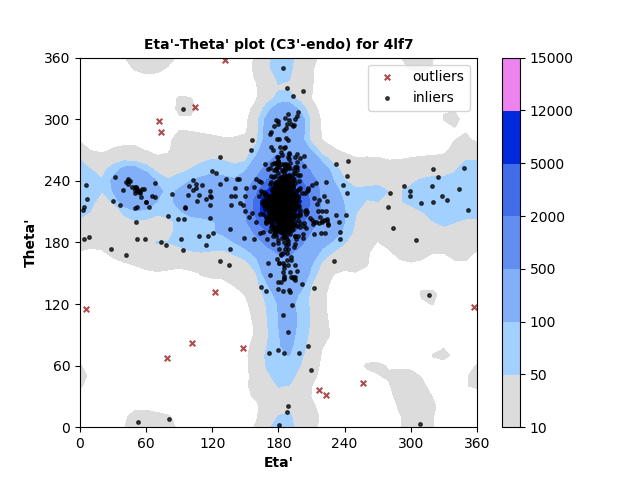
1310 C3'-endo RNA residues with 13 outliers ( 0.99 % ) from Eta'-Theta' plot
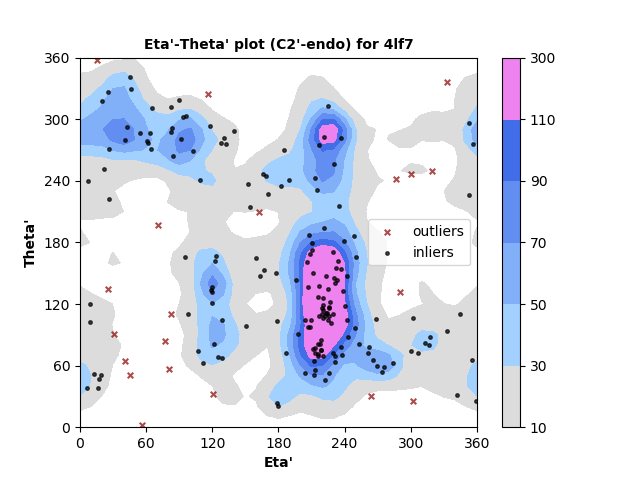
193 C2'-endo RNA residues with 20 outliers ( 10.36 % ) from Eta'-Theta' plot
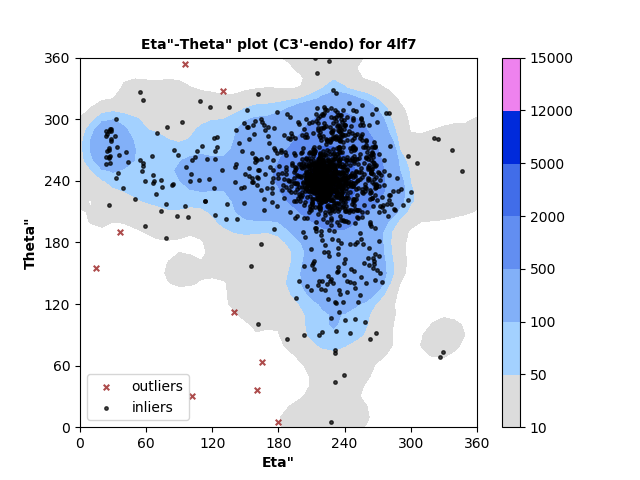
1310 C3'-endo RNA residues with 9 outliers ( 0.69 % ) from Eta"-Theta" plot
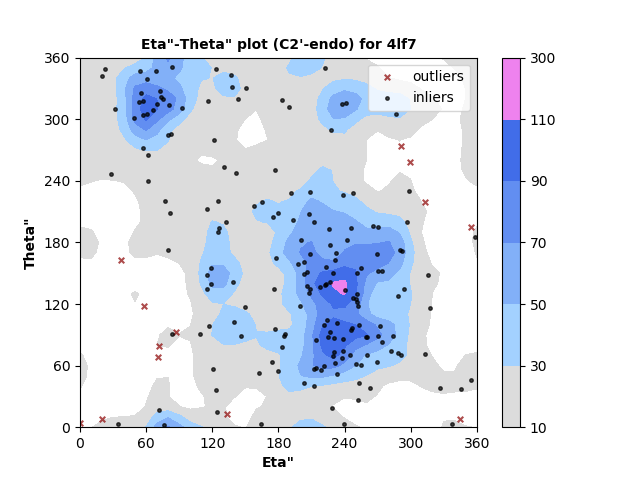
193 C2'-endo RNA residues with 13 outliers ( 6.74 % ) from Eta"-Theta" plot
List of Outliers
| Model Number | Chain | Residue Type | Residue Identifier | Eta | Theta | Eta' | Theta' | Eta'' | Theta'' | Sugar Pucker | Suiteness |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | U | 204 | 228.2 | 80.0 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 247 | 263.7 | 132.6 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 352 | 83.3 | 91.9 | 79.3 | 67.6 | 95.2 | 354.1 | C3'endo | 0.0 |
| 1 | A | A | 353 | 11.7 | 110.5 | 5.8 | 115.2 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 530 | 204.5 | 35.7 | 217.2 | 36.5 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 563 | 107.3 | 106.2 | 101.7 | 82.4 | 101.5 | 30.4 | C3'endo | 0.105 |
| 1 | A | U | 723 | 70.6 | 289.7 | 73.4 | 287.2 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 754 | 115.8 | 306.0 | 104.7 | 311.7 | --- | --- | C3'endo | 0.649 |
| 1 | A | U | 839 | 94.4 | 308.0 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 841 | 26.9 | 315.5 | 71.7 | 298.6 | 129.7 | 327.8 | C3'endo | 0.0 |
| 1 | A | C | 1054 | 18.7 | 117.2 | 357.9 | 116.8 | 35.9 | 189.7 | C3'endo | 0.014 |
| 1 | A | C | 1200 | 219.9 | 61.9 | 222.7 | 31.7 | --- | --- | C3'endo | 0.01 |
| 1 | A | U | 1278 | 245.8 | 45.9 | 257.0 | 42.8 | --- | --- | C3'endo | 0.528 |
| 1 | A | A | 1398 | 128.6 | 49.3 | 147.8 | 77.2 | --- | --- | C3'endo | 0.01 |
| 1 | A | A | 1446 | 4.6 | 165.9 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1530 | 130.7 | 110.2 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 722 | --- | --- | 122.6 | 131.9 | 165.4 | 63.3 | C3'endo | 0.871 |
| 1 | A | C | 1362 | --- | --- | 131.4 | 357.6 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 6 | --- | --- | --- | --- | 139.4 | 111.9 | C3'endo | 0.354 |
| 1 | A | A | 665 | --- | --- | --- | --- | 14.4 | 154.7 | C3'endo | 0.0 |
| 1 | A | G | 1442 | --- | --- | --- | --- | 160.3 | 36.5 | C3'endo | 0.0 |
| 1 | A | G | 1505 | --- | --- | --- | --- | 179.5 | 5.3 | C3'endo | 0.034 |
| 1 | A | A | 51 | 24.8 | 49.0 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 130 | 357.7 | 254.6 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | G | 190F | 42.9 | 347.5 | 116.3 | 324.8 | --- | --- | C2'endo | 0.087 |
| 1 | A | A | 532 | 351.2 | 254.2 | 300.5 | 246.3 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 533 | 261.3 | 292.7 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 815 | 68.6 | 20.9 | 56.3 | 2.4 | --- | --- | C2'endo | 0.16 |
| 1 | A | A | 819 | 299.0 | 304.5 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 820 | 171.0 | 6.9 | --- | --- | --- | --- | C2'endo | 0.488 |
| 1 | A | A | 974 | 78.6 | 59.7 | 121.0 | 32.6 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 975 | 39.3 | 60.6 | 41.2 | 65.0 | 70.9 | 68.7 | C2'endo | 0.0 |
| 1 | A | C | 1027 | 134.9 | 184.3 | 162.4 | 209.5 | --- | --- | C2'endo | 0.0 |
| 1 | A | G | 1094 | 20.4 | 58.7 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | C | 1129 | 316.9 | 134.6 | 290.1 | 131.9 | --- | --- | C2'endo | 0.0 |
| 1 | A | G | 1443 | 55.4 | 63.8 | 45.3 | 50.6 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 8 | --- | --- | 287.0 | 241.3 | 299.4 | 258.1 | C2'endo | 0.222 |
| 1 | A | A | 50 | --- | --- | 76.9 | 83.5 | --- | --- | C2'endo | 0.257 |
| 1 | A | A | 60 | --- | --- | 25.2 | 135.0 | --- | --- | C2'endo | 0.478 |
| 1 | A | U | 190E | --- | --- | 319.0 | 249.7 | 313.4 | 219.1 | C2'endo | 0.059 |
| 1 | A | A | 451 | --- | --- | 82.3 | 110.5 | --- | --- | C2'endo | 0.093 |
| 1 | A | A | 460 | --- | --- | 71.0 | 197.2 | 36.9 | 162.7 | C2'endo | 0.0 |
| 1 | A | U | 960 | --- | --- | 263.8 | 30.7 | 291.6 | 273.6 | C2'endo | 0.0 |
| 1 | A | U | 1125 | --- | --- | 15.7 | 357.8 | 20.2 | 7.5 | C2'endo | 0.0 |
| 1 | A | U | 1196 | --- | --- | 301.9 | 25.2 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1201 | --- | --- | 31.1 | 90.9 | 87.5 | 92.8 | C2'endo | 0.0 |
| 1 | A | U | 1257 | --- | --- | 333.0 | 335.8 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1363 | --- | --- | 81.2 | 56.6 | 133.1 | 12.8 | C2'endo | 0.01 |
| 1 | A | U | 65 | --- | --- | --- | --- | 0.3 | 3.7 | C2'endo | 0.889 |
| 1 | A | G | 129A | --- | --- | --- | --- | 345.1 | 8.4 | C2'endo | 0.0 |
| 1 | A | U | 561 | --- | --- | --- | --- | 58.4 | 118.4 | C2'endo | 0.59 |
| 1 | A | A | 1157 | --- | --- | --- | --- | 71.3 | 79.3 | C2'endo | 0.18 |
| 1 | A | A | 1183 | --- | --- | --- | --- | 354.7 | 194.8 | C2'endo | 0.804 |