RNA Pseudotorsion Plots for PDB entry 4x62
Crystal Structure of 30S ribosomal subunit from Thermus thermophilus Deposited 2014-12-06 X-RAY DIFFRACTION resol 3.4492 Å
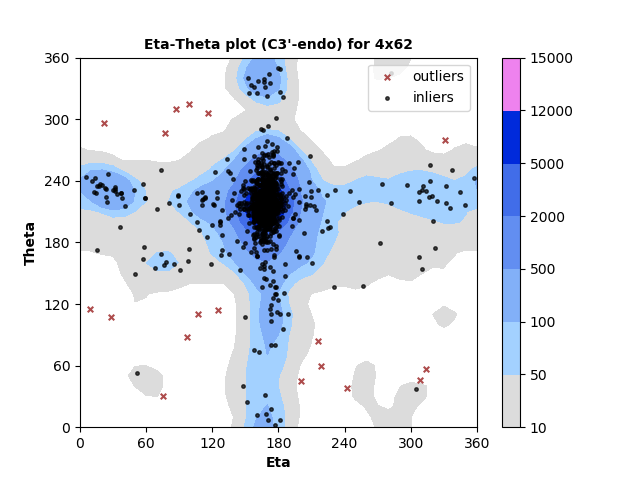
1333 C3'-endo RNA residues with 18 outliers ( 1.35 % ) from Eta-Theta plot
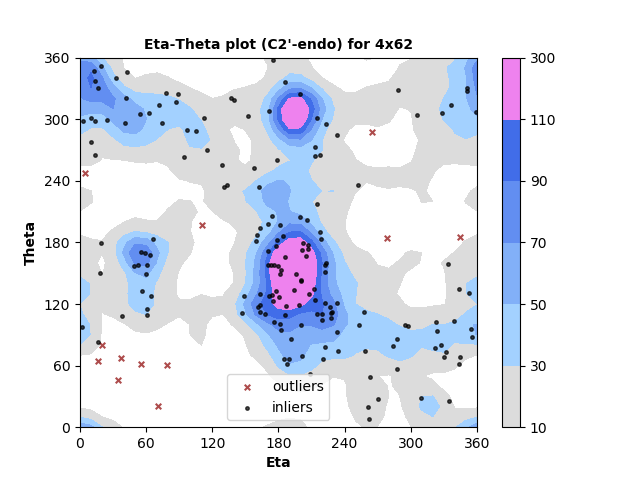
185 C2'-endo RNA residues with 12 outliers ( 6.49 % ) from Eta-Theta plot
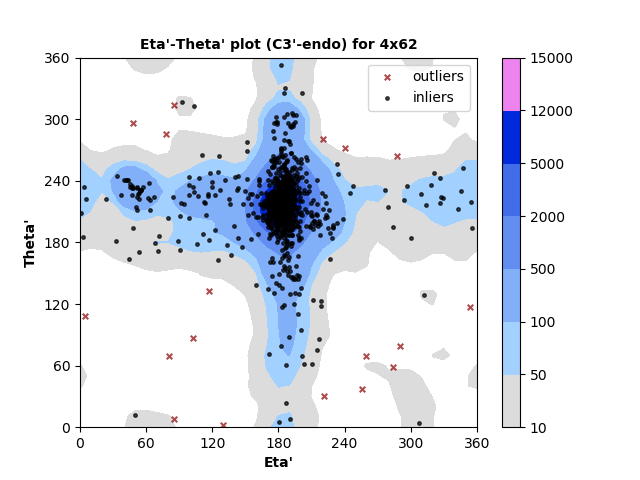
1333 C3'-endo RNA residues with 18 outliers ( 1.35 % ) from Eta'-Theta' plot
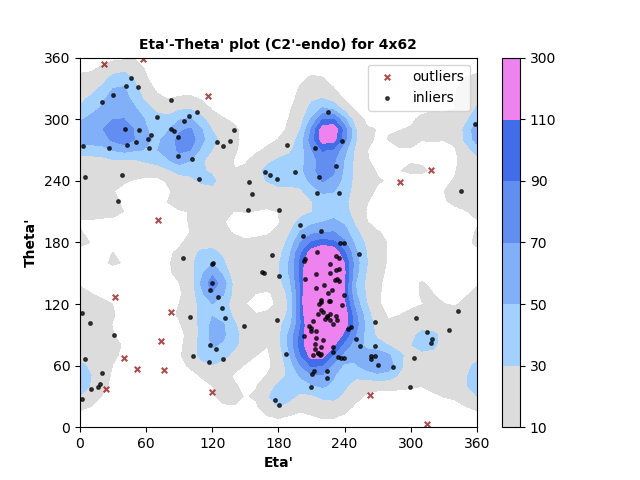
185 C2'-endo RNA residues with 16 outliers ( 8.65 % ) from Eta'-Theta' plot
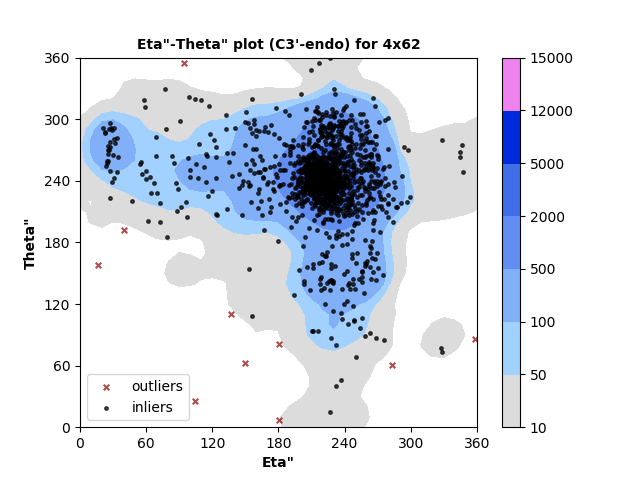
1333 C3'-endo RNA residues with 10 outliers ( 0.75 % ) from Eta"-Theta" plot
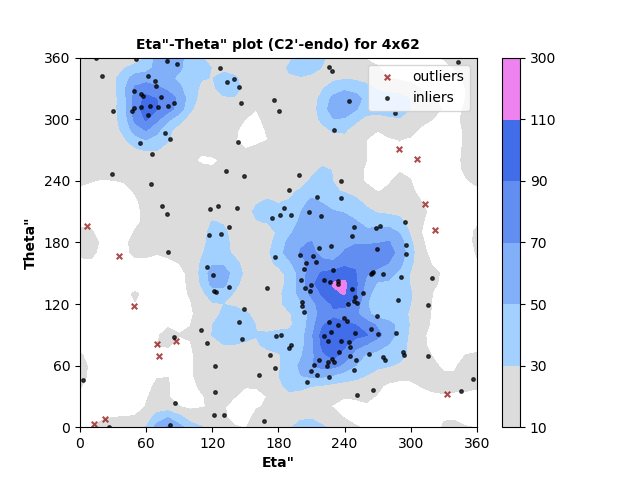
185 C2'-endo RNA residues with 13 outliers ( 7.03 % ) from Eta"-Theta" plot
List of Outliers
| Model Number | Chain | Residue Type | Residue Identifier | Eta | Theta | Eta' | Theta' | Eta'' | Theta'' | Sugar Pucker | Suiteness |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | U | 204 | 215.6 | 83.7 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 352 | 97.1 | 87.7 | 80.4 | 69.2 | 94.6 | 354.8 | C3'endo | 0.0 |
| 1 | A | A | 353 | 8.8 | 115.2 | 4.9 | 108.0 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 530 | 200.4 | 44.5 | --- | --- | --- | --- | C3'endo | 0.362 |
| 1 | A | A | 532 | 330.9 | 279.4 | 287.6 | 264.4 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 563 | 106.7 | 110.2 | 102.3 | 86.9 | 104.0 | 25.6 | C3'endo | 0.01 |
| 1 | A | U | 723 | 77.1 | 286.4 | 78.3 | 285.9 | --- | --- | C3'endo | 0.01 |
| 1 | A | C | 754 | 116.4 | 306.2 | --- | --- | --- | --- | C3'endo | 0.9 |
| 1 | A | A | 816 | 75.1 | 30.5 | 85.3 | 8.3 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 839 | 86.9 | 310.3 | 85.0 | 314.1 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 841 | 21.7 | 296.6 | 47.8 | 295.9 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1054 | 28.5 | 107.6 | 354.1 | 116.7 | 358.5 | 85.5 | C3'endo | 0.0 |
| 1 | A | U | 1136 | 98.7 | 314.3 | --- | --- | --- | --- | C3'endo | 0.01 |
| 1 | A | C | 1200 | 218.4 | 59.9 | 221.0 | 30.7 | --- | --- | C3'endo | 0.01 |
| 1 | A | U | 1212 | 313.9 | 57.1 | 283.6 | 58.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1278 | 242.5 | 37.8 | 255.7 | 37.1 | --- | --- | C3'endo | 0.512 |
| 1 | A | G | 1300 | 308.8 | 46.4 | 290.3 | 78.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1530 | 125.1 | 114.2 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 722 | --- | --- | 117.3 | 133.0 | 150.0 | 62.3 | C3'endo | 0.166 |
| 1 | A | A | 1005 | --- | --- | 240.5 | 272.0 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1281 | --- | --- | 220.9 | 281.2 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1347 | --- | --- | 259.8 | 69.7 | --- | --- | C3'endo | 0.046 |
| 1 | A | C | 1362 | --- | --- | 129.5 | 1.7 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 6 | --- | --- | --- | --- | 137.0 | 110.5 | C3'endo | 0.349 |
| 1 | A | A | 665 | --- | --- | --- | --- | 16.2 | 157.9 | C3'endo | 0.0 |
| 1 | A | G | 1182 | --- | --- | --- | --- | 283.0 | 60.5 | C3'endo | 0.32 |
| 1 | A | A | 1377 | --- | --- | --- | --- | 180.7 | 80.6 | C3'endo | 0.863 |
| 1 | A | A | 1492 | --- | --- | --- | --- | 39.5 | 191.9 | C3'endo | 0.0 |
| 1 | A | G | 1505 | --- | --- | --- | --- | 180.9 | 7.5 | C3'endo | 0.0 |
| 1 | A | A | 51 | 34.1 | 45.6 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 60 | 278.4 | 184.2 | 31.8 | 126.6 | --- | --- | C2'endo | 0.116 |
| 1 | A | A | 130 | 4.2 | 248.0 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 244 | 19.9 | 79.8 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | C | 748 | 110.4 | 196.5 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 815 | 70.6 | 20.2 | 57.2 | 359.1 | --- | --- | C2'endo | 0.01 |
| 1 | A | A | 974 | 78.8 | 60.6 | 119.5 | 33.8 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 975 | 37.3 | 67.6 | 40.1 | 67.2 | 71.3 | 69.5 | C2'endo | 0.0 |
| 1 | A | G | 1094 | 16.3 | 64.0 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1285 | 344.8 | 185.5 | --- | --- | --- | --- | C2'endo | 0.321 |
| 1 | A | G | 1443 | 55.5 | 61.5 | 52.1 | 56.9 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 8 | --- | --- | 290.5 | 238.6 | 305.7 | 261.5 | C2'endo | 0.255 |
| 1 | A | A | 50 | --- | --- | 73.1 | 83.8 | --- | --- | C2'endo | 0.084 |
| 1 | A | G | 129A | --- | --- | 23.8 | 37.5 | 12.3 | 2.7 | C2'endo | 0.0 |
| 1 | A | U | 190E | --- | --- | 318.8 | 250.7 | 313.2 | 217.8 | C2'endo | 0.01 |
| 1 | A | G | 190F | --- | --- | 115.9 | 323.0 | --- | --- | C2'endo | 0.039 |
| 1 | A | A | 451 | --- | --- | 82.6 | 112.1 | --- | --- | C2'endo | 0.496 |
| 1 | A | A | 460 | --- | --- | 70.4 | 201.4 | 35.1 | 167.0 | C2'endo | 0.0 |
| 1 | A | U | 960 | --- | --- | 262.8 | 31.4 | 289.1 | 270.7 | C2'endo | 0.0 |
| 1 | A | U | 1125 | --- | --- | 21.3 | 354.0 | --- | --- | C2'endo | 0.0 |
| 1 | A | U | 1257 | --- | --- | 315.2 | 3.5 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1363 | --- | --- | 76.1 | 55.3 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 533 | 264.5 | 287.2 | --- | --- | 321.8 | 191.9 | C2'endo | 0.0 |
| 1 | A | U | 561 | --- | --- | --- | --- | 48.8 | 117.9 | C2'endo | 0.398 |
| 1 | A | U | 1049 | --- | --- | --- | --- | 332.7 | 32.2 | C2'endo | 0.264 |
| 1 | A | A | 1157 | --- | --- | --- | --- | 69.9 | 81.3 | C2'endo | 0.1 |
| 1 | A | A | 1183 | --- | --- | --- | --- | 6.6 | 196.1 | C2'endo | 0.0 |
| 1 | A | A | 1201 | --- | --- | --- | --- | 86.8 | 84.1 | C2'endo | 0.0 |
| 1 | A | U | 1240 | --- | --- | --- | --- | 22.4 | 7.6 | C2'endo | 0.902 |