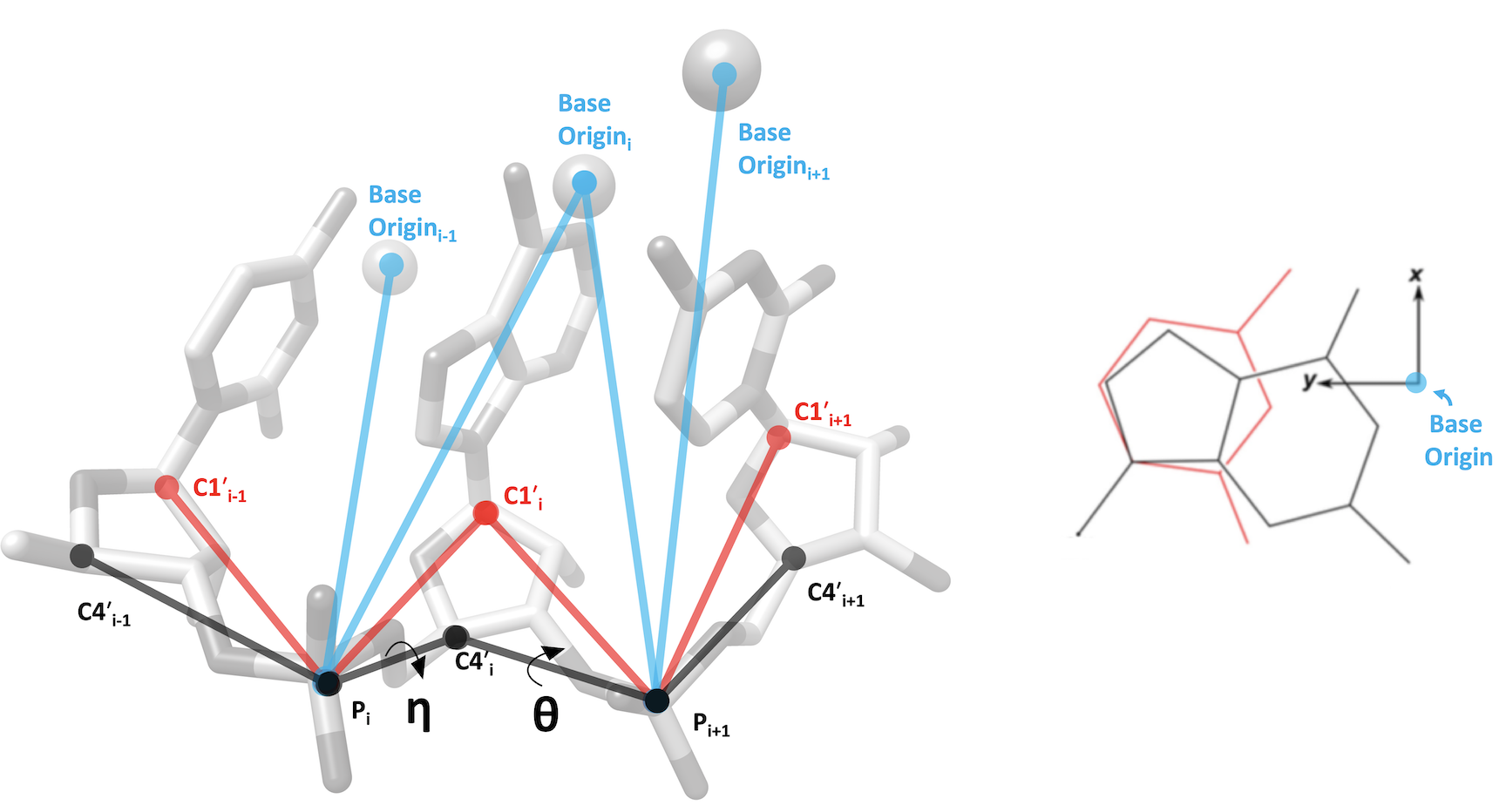
RNA Pseudotorsion Plots for PDB entry 6mpf
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGC-codon in the A-site and paromomycin Deposited 2018-10-05 X-RAY DIFFRACTION resol 3.33 Å
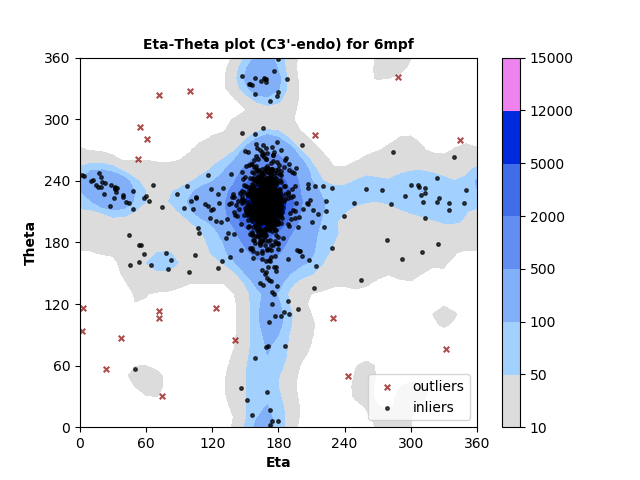
1318 C3'-endo RNA residues with 21 outliers ( 1.59 % ) from Eta-Theta plot
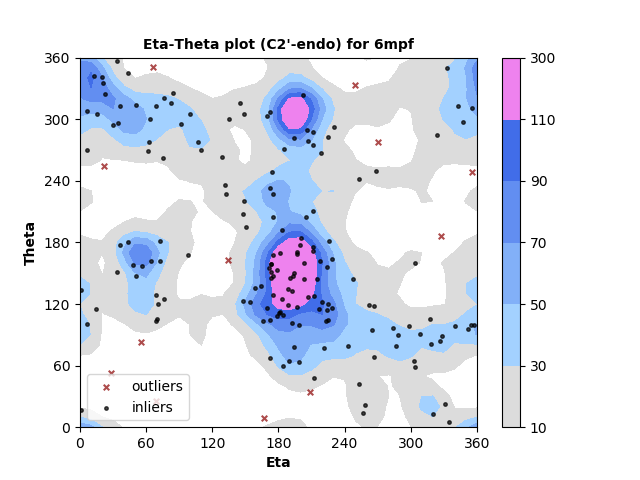
179 C2'-endo RNA residues with 12 outliers ( 6.7 % ) from Eta-Theta plot
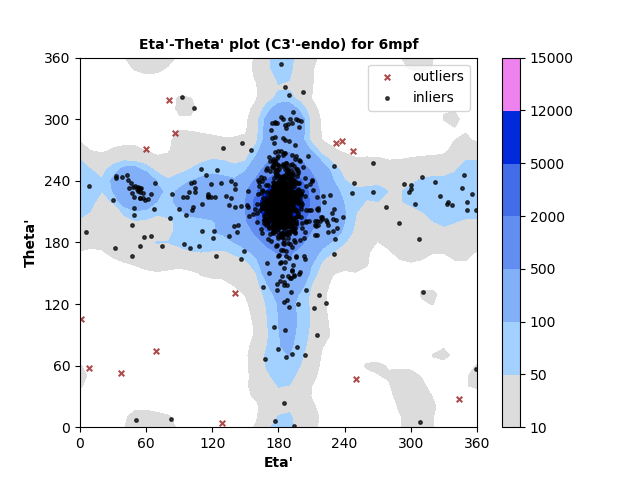
1318 C3'-endo RNA residues with 14 outliers ( 1.06 % ) from Eta'-Theta' plot
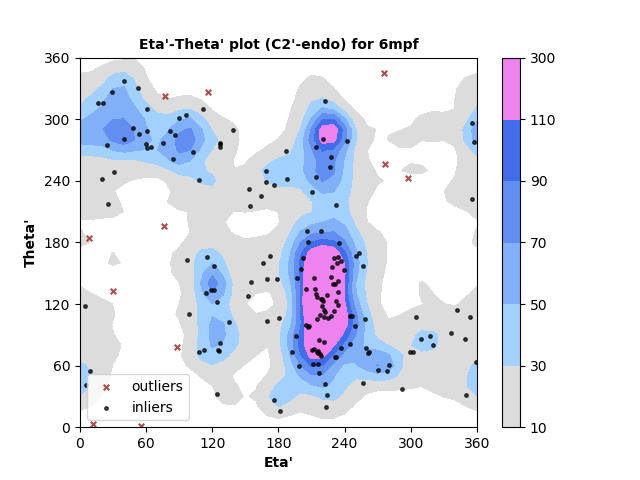
179 C2'-endo RNA residues with 11 outliers ( 6.15 % ) from Eta'-Theta' plot
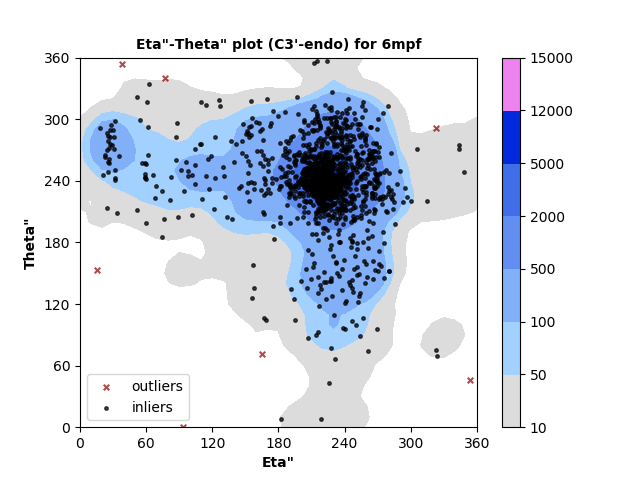
1318 C3'-endo RNA residues with 7 outliers ( 0.53 % ) from Eta"-Theta" plot
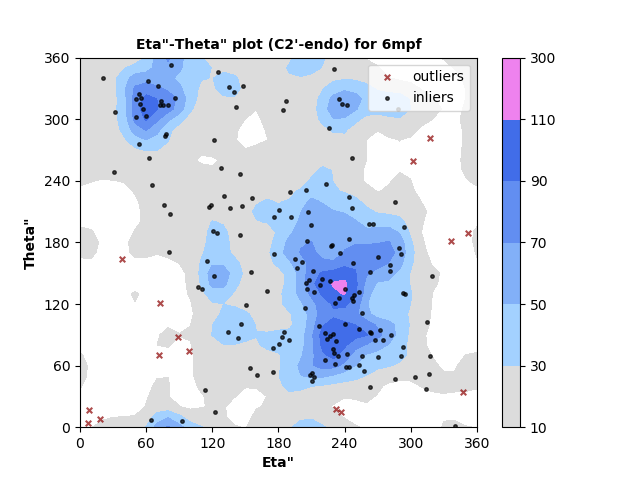
179 C2'-endo RNA residues with 15 outliers ( 8.38 % ) from Eta"-Theta" plot
List of Outliers
| Model Number | Chain | Residue Type | Residue Identifier | Eta | Theta | Eta' | Theta' | Eta'' | Theta'' | Sugar Pucker | Suiteness |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | U | 210 | 229.7 | 106.2 | --- | --- | --- | --- | C3'endo | 0.377 |
| 1 | A | C | 347 | 71.5 | 106.5 | 69.4 | 74.1 | 93.3 | 0.4 | C3'endo | 0.0 |
| 1 | A | A | 348 | 3.1 | 116.5 | 0.8 | 105.1 | --- | --- | C3'endo | 0.01 |
| 1 | A | U | 363 | 288.4 | 341.4 | --- | --- | --- | --- | C3'endo | 0.275 |
| 1 | A | U | 543 | 2.2 | 93.3 | --- | --- | --- | --- | C3'endo | 0.17 |
| 1 | A | U | 706 | 60.7 | 280.8 | 59.8 | 270.6 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 737 | 116.7 | 303.7 | --- | --- | --- | --- | C3'endo | 0.734 |
| 1 | A | A | 799 | 74.2 | 30.5 | --- | --- | --- | --- | C3'endo | 0.431 |
| 1 | A | U | 822 | 71.4 | 323.4 | 80.8 | 318.3 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 824 | 54.5 | 292.1 | 86.2 | 286.4 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1076 | 23.9 | 56.6 | 8.3 | 58.0 | 353.6 | 46.0 | C3'endo | 0.0 |
| 1 | A | U | 1107 | 332.2 | 76.2 | 344.1 | 27.1 | 37.7 | 353.8 | C3'endo | 0.0 |
| 1 | A | U | 1108 | 52.3 | 261.4 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1112 | 213.4 | 284.8 | 237.6 | 278.8 | --- | --- | C3'endo | 0.0 |
| 1 | A | U | 1259 | 243.5 | 49.9 | 250.4 | 47.2 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1380 | 140.4 | 85.1 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1425 | 37.0 | 86.9 | 36.8 | 52.5 | --- | --- | C3'endo | 0.0 |
| 1 | A | G | 1507 | 123.8 | 115.6 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1510 | 344.6 | 279.5 | --- | --- | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1511 | 72.1 | 113.5 | 140.4 | 130.4 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 983 | --- | --- | 247.5 | 269.0 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 1206 | --- | --- | 232.3 | 276.4 | --- | --- | C3'endo | 0.0 |
| 1 | A | C | 1344 | --- | --- | 129.0 | 3.9 | --- | --- | C3'endo | 0.0 |
| 1 | A | A | 648 | --- | --- | --- | --- | 15.4 | 153.0 | C3'endo | 0.0 |
| 1 | A | U | 1118 | 99.6 | 327.3 | --- | --- | 76.7 | 340.3 | C3'endo | 0.02 |
| 1 | A | G | 1183 | --- | --- | --- | --- | 322.9 | 291.1 | C3'endo | 0.0 |
| 1 | A | A | 1359 | --- | --- | --- | --- | 165.4 | 70.9 | C3'endo | 0.627 |
| 1 | A | A | 124 | 355.7 | 248.9 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | G | 211 | 134.2 | 162.5 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 339 | 66.4 | 350.6 | 77.5 | 322.2 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 454 | 21.9 | 254.7 | 76.2 | 195.6 | 37.7 | 163.6 | C2'endo | 0.0 |
| 1 | A | G | 513 | 208.5 | 34.1 | --- | --- | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 798 | 69.2 | 25.9 | 55.5 | 1.6 | --- | --- | C2'endo | 0.027 |
| 1 | A | U | 803 | 167.3 | 9.4 | --- | --- | --- | --- | C2'endo | 0.535 |
| 1 | A | A | 951 | 55.7 | 83.2 | --- | --- | --- | --- | C2'endo | 0.222 |
| 1 | A | C | 1111 | 249.7 | 333.7 | 275.8 | 344.9 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 1426 | 327.4 | 186.6 | 8.5 | 184.4 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 8 | --- | --- | 297.4 | 242.7 | 302.0 | 259.0 | C2'endo | 0.121 |
| 1 | A | A | 50 | --- | --- | 88.5 | 78.0 | 89.2 | 87.6 | C2'endo | 0.0 |
| 1 | A | A | 60 | --- | --- | 30.3 | 132.2 | --- | --- | C2'endo | 0.364 |
| 1 | A | G | 190 | --- | --- | 116.3 | 327.0 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 982 | --- | --- | 276.7 | 256.7 | 317.5 | 282.0 | C2'endo | 0.0 |
| 1 | A | C | 1379 | --- | --- | 12.1 | 2.8 | --- | --- | C2'endo | 0.0 |
| 1 | A | A | 51 | 28.3 | 53.0 | --- | --- | 347.8 | 34.1 | C2'endo | 0.0 |
| 1 | A | U | 65 | --- | --- | --- | --- | 8.3 | 17.2 | C2'endo | 0.0 |
| 1 | A | A | 516 | 270.3 | 277.6 | --- | --- | 336.3 | 181.3 | C2'endo | 0.0 |
| 1 | A | U | 544 | --- | --- | --- | --- | 72.6 | 120.8 | C2'endo | 0.0 |
| 1 | A | G | 704 | --- | --- | --- | --- | 98.9 | 74.4 | C2'endo | 0.0 |
| 1 | A | C | 823 | --- | --- | --- | --- | 237.0 | 14.4 | C2'endo | 0.0 |
| 1 | A | A | 1139 | --- | --- | --- | --- | 71.8 | 69.9 | C2'endo | 0.0 |
| 1 | A | A | 1164 | --- | --- | --- | --- | 352.1 | 189.1 | C2'endo | 0.0 |
| 1 | A | U | 1221 | --- | --- | --- | --- | 18.0 | 8.3 | C2'endo | 0.0 |
| 1 | A | U | 1238 | --- | --- | --- | --- | 232.2 | 17.3 | C2'endo | 0.0 |
| 1 | A | C | 1382 | --- | --- | --- | --- | 7.3 | 3.9 | C2'endo | 0.0 |